News Center
Detecting short tandem repeats using WGS
- By Variantyx, Inc
- Posted in Clinical Education
In this week’s post we’re taking a closer look at short tandem repeats (STRs) and how they’re detected using whole genome sequencing (WGS).
As their name suggests, STR’s are short sequences of DNA, typically 1 to 6 nucleotides in length, that repeat consecutively. The number of repeats varies from person to person, with the length at times expanding during transmission from parent to offspring. Most repeats do not have a discernible function, but some have the potential to become pathogenic when the number of repeats exceeds a particular threshold. For example, in previous posts, we’ve talked about the role of repeat expansions in fragile X syndrome and inherited ataxias.
To recap, fragile X syndrome is caused by expansion of the unstable CGG repeat within the 5’ UTR of the FMR1 gene. Individuals with alleles less than 40 repeats in length are unaffected, those with premutation alleles up to 200 repeats in length are at risk of passing an expanded pathogenic allele on to their offspring and those with pathogenic alleles of more than 200 repeats typically exhibit symptoms of the disease.
Inherited ataxias come in multiple forms and have multiple causes. Friedreich’s ataxia is caused by expansion of the GAA repeat within the FXN gene. In contrast, spinocerebellar ataxia can be caused by expansion of the CAG repeat within a number of different genes including ATXN1, ATXN2, ATXN3 and numerous others.
The ability to detect repeat expansions and determine the count of repeats within individual alleles is an important part of the rare disease diagnostic process. Until recently, detecting repeat expansions has required the use of PCR or southern blot analysis, usually employed to interrogate a single targeted gene. With the introduction of clinical WGS it is now possible to screen the full genome for pathogenic repeat expansions, when paired with the right algorithms.
At Variantyx, our algorithms use three separate paired-end read strategies to detect repeat expansion alleles in more than twenty known pathogenic loci, all within a single assay.
The first strategy
The first strategy focuses on short range variants that are less than 50 repeats in length. Here de novo assembly of spanning reads is used as the full length of the repeat is contained within either R1 or R2, with uniquely mappable flanking sequences.

The second strategy
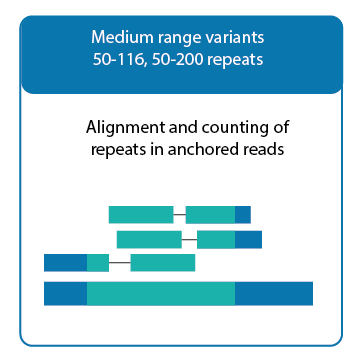
The second strategy focuses on medium range variants that range from 50 to 116 or 50 to 200 repeats in length, with the upper limit determined by whether the sequencing insert size is 350 bp or 550 bp. Here alignment and counting of repeats in anchored reads is used. With anchored reads, one member of the pair, either R1 or R2, contains only repeat sequence while the other member contains partial repeat sequence and partial uniquely mappable sequence.
The final strategy
The final strategy focuses on long range variants that are >116 or >200 repeats in length. Here counting and statistical normalization of reads containing only repeat sequence is used to estimate the repeat length.

Combing the three different methods, repeat length is calculated with good specificity up to the threshold that is determined by the sequencing insert size.
View the specific tandem repeat expansion genes and disorders tested by our algorithms.
