
MISSED BY OTHERS, DETECTED BY US
Genomic Unity® Case Study
Clinical presentation
A 42-year-old male presented with multiple symptoms including:
- Progressive dysphagia
- Memory impairment
- Ambulatory dysfunction
- Body twitches
Previous genetic testing
A lysosomal storage disorders enzyme panel test was negative. Additional targeted genetic tests performed based on suspected clinical diagnoses were negative including:
- HTT testing
- MERFF targeted variant testing

Genomic Unity® Testing
was ordered because of its ability to identify all major variant types in a single test.
Results and interpretation
Variantyx Genomic Unity® testing identified likely compound heterozygous variants in the VPS13A gene: a likely pathogenic single nucleotide deletion that results in frameshift and a likely pathogenic AluY mobile element insertion (MEI) that disrupts exon 33.
Diagnosis: Chorea-acanthocytosis
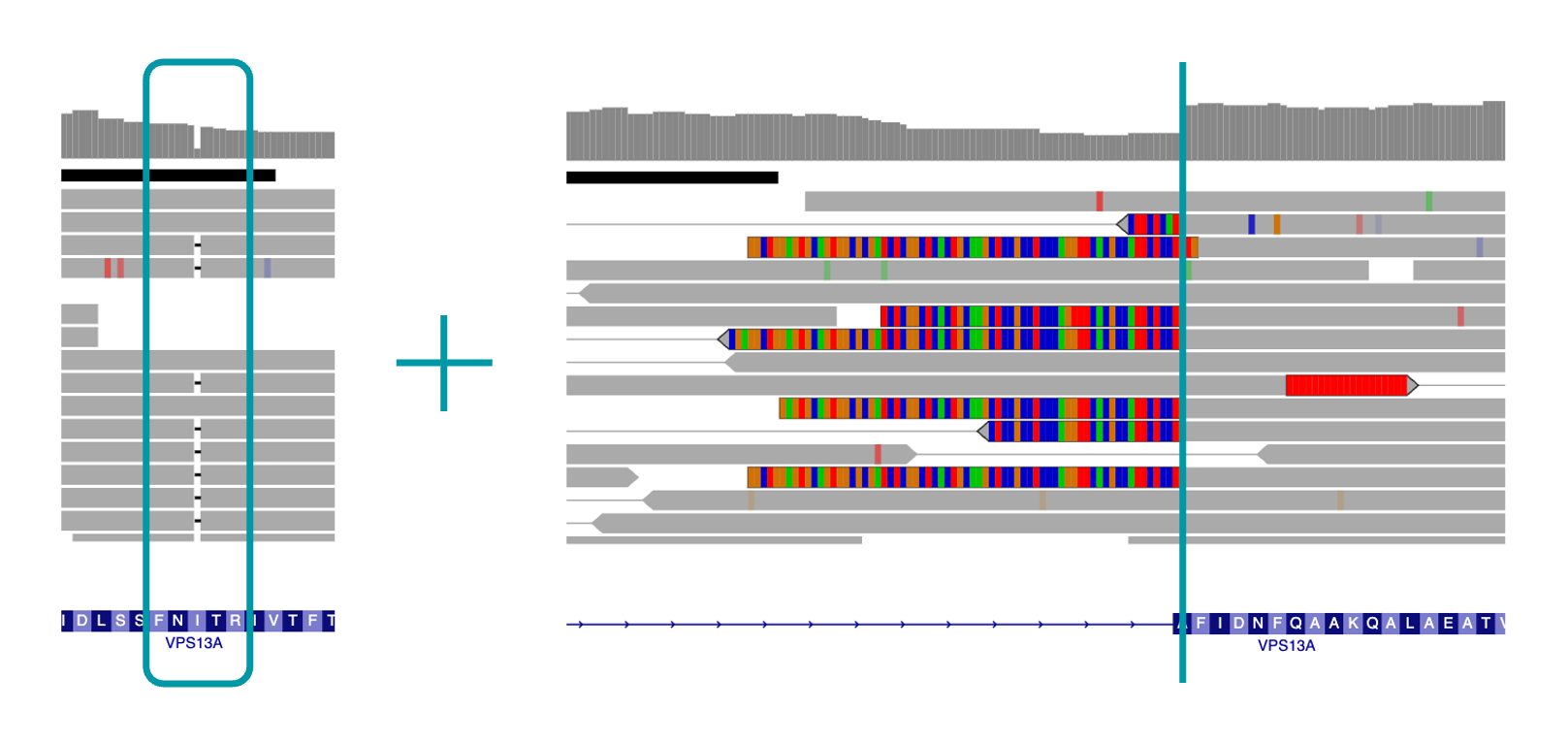
Uniform data from WGS clearly shows both the single nucleotide deletion (left) and the MEI breakpoint (right).
The Variantyx Difference
Why were these likely compound heterozygous variants detected by Genomic Unity® testing, and not detected by other tests?
-
The VPS13A gene was not included in the targeted gene and variant testing performed, and would be unlikely to be selected based on the clinical presentation.
Variantyx genome analysis does not exclude any gene.
-
Mobile element insertions (MEI) are undetectable by most available technologies – including gene sequencing, panel and exome tests.
Variantyx genome analysis detects many types of structural variants including copy number variants, deletions/duplications, inversions, mobile element insertions, regions of homozygosity and aneuploidy.
Variantyx tests that would have identified these variants
Want similar results for your patients?
Connect with a Clinical Specialist to find out how easy it is to bring the power of whole genome sequencing into your practice.


