MISSED BY OTHERS, DETECTED BY US
Genomic Unity® Case Study
Clinical presentation
A 10-year-old female presented with a history of developmental delays and intellectual disability. Additional symptoms included:
- Seizures
- Generalized hypotonia
- Hypercholesterolemia
- Sleep disturbance
Previous genetic testing
Extensive prior genetic testing was performed with negative results, including:
- Chromosomal microarray (x2)
- FMR1 testing
- Prader-Willi and Angelman syndrome testing
- Mitochondrial genome sequencing
- Whole exome sequencing with reanalysis (x2)
Genomic Unity® Testing
was ordered because of its ability to identify all major variant types in a single test.
Results and interpretation
Variantyx Genomic Unity® testing identified a de novo, heterozygous, likely pathogenic single nucleotide variant in the SYNGAP1 gene that results in a p.Glu221Val change.
Diagnosis: SYNGAP1-related developmental disorders
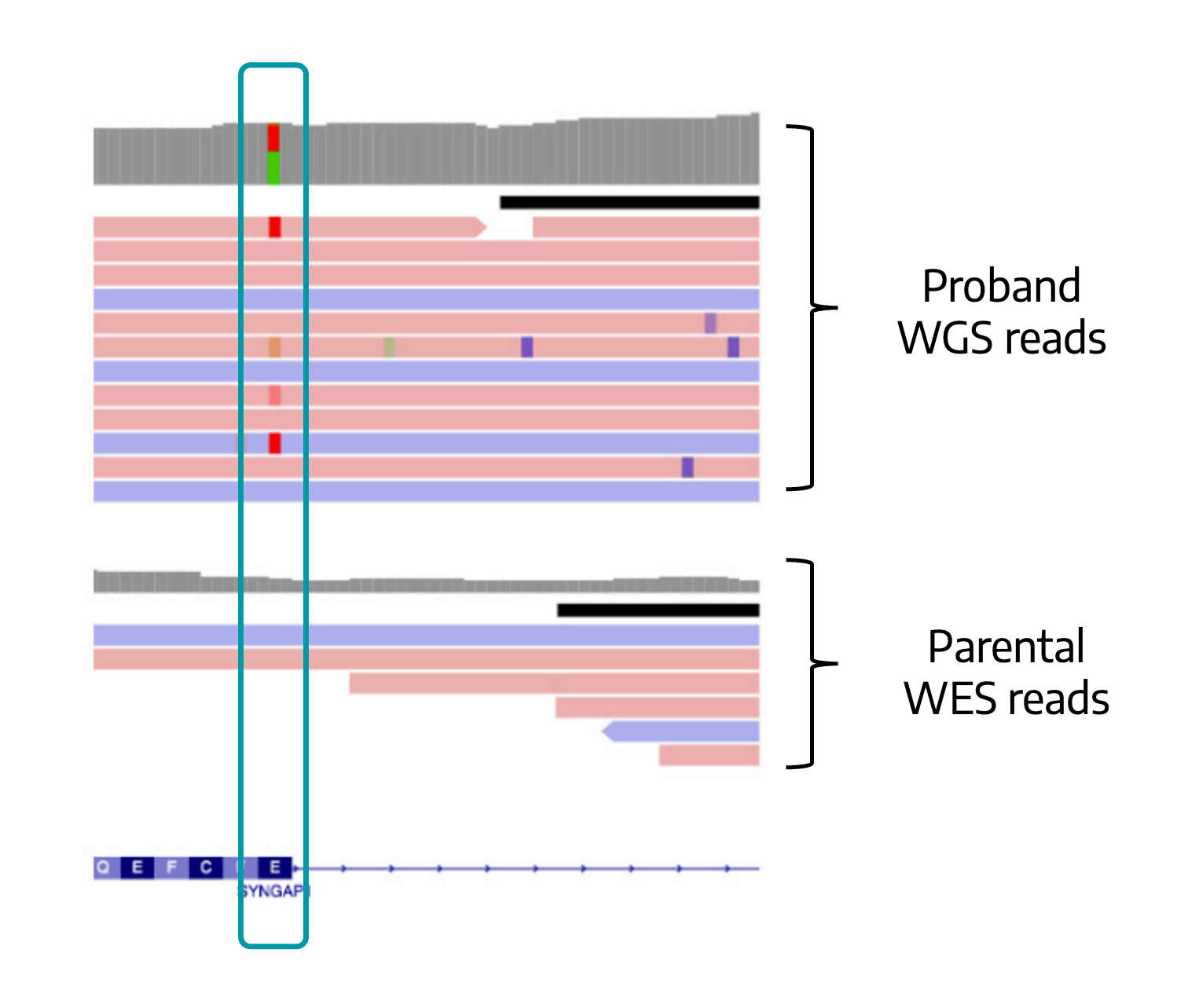
Uniform data from WGS clearly shows a heterozygous variant that falls within a region of poor WES coverage.
The Variantyx Difference
Why was this variant detected by Genomic Unity® testing, and not detected by prior testing – including exome testing?
-
Small sequence changes can’t be detected by CMA.
-
While sequence changes are usually detectable by exome testing, this variant is located near an exon/intron boundary, in a region that is poorly covered by WES probes. This results in poor read coverage.
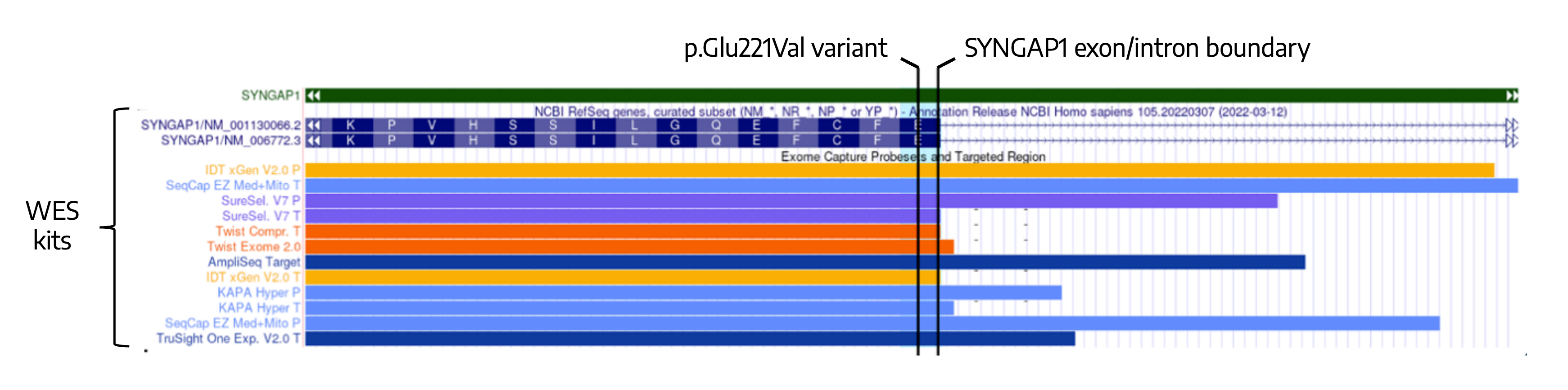
Variantyx genome sequencing does not use PCR amplification. Its uniform coverage depth across the entire gene makes it easy to identify this exonic single nucleotide change. Changes in other genes are simultaneously ruled out without the need for gene-specific tests like FMR1 that extend the diagnostic odyssey.
Variantyx tests that would have identified this variant
Want similar results for your patients?
Connect with a Clinical Specialist to find out how easy it is to bring the power of whole genome sequencing into your practice.
