
MISSED BY OTHERS, DETECTED BY US
Genomic Unity® Case Study
Clinical presentation
A 16-year-old male had been denied access to DMD trials and vamorolone treatment due to lack of a molecular diagnosis, despite presenting with a clinical diagnosis of Duchenne muscular dystrophy based on muscle biopsy results and hallmark symptoms:
- Motor developmental delays, large calves, toe walking, frequent falls, loss of independent ambulation at age 10
- Elevated CK levels
- Restrictive lung disease
Previous genetic testing
Multiple tests were performed with negative results including:
- DMD gene sequencing (x2)
- DMD del/dup analysis (x2)
- Dystroglycan-related congenital muscular dystrophy panel

Genomic Unity® Testing
was ordered because of its ability to identify all major variant types in a single test.
Results and interpretation
Variantyx Genomic Unity® testing identified a hemizygous, pathogenic 713 kb inversion encompassing exons 45-57 of the DMD gene.
The inversion is expected to result in loss of function which is consistent with prior RNA analysis identifying an intronal inclusion that leads to an inability to produce dystrophin.
Diagnosis: Duchenne muscular dystrophy
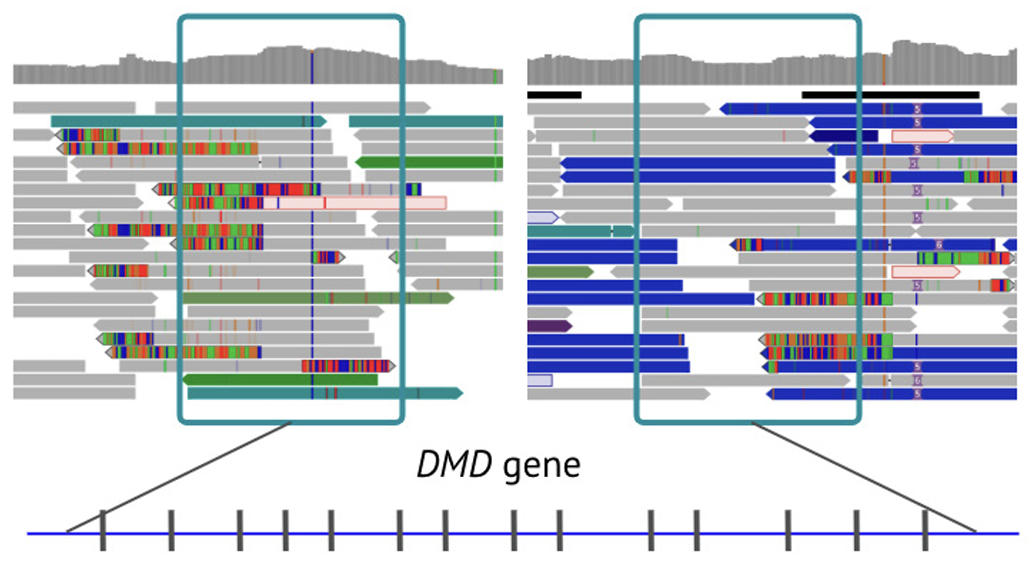
Uniform data from WGS identifies the breakpoints of the 713 kb inversion spanning DMD exons 45-57.
The Variantyx Difference
Why was this inversion detected by Genomic Unity® testing, and not detected by other targeted DMD tests?
-
Balanced rearrangements like inversions are undetectable by most available technologies – including MLPA and targeted gene sequencing.
Variantyx genome analysis detects many types of structural variants including copy number variants, deletions/duplications, inversions, mobile element insertions, regions of homozygosity and aneuploidy. -
Both inversion breakpoints are intronic, adding to the complexity of detection.
Variantyx genome analysis includes intronic regions, enabling breakpoint detection regardless of location.
Variantyx tests that would have identified this variant
Genomic Unity® 2.0 | Genomic Unity® Whole Genome Analysis | Genomic Unity® Lightning Genome Analysis | Genomic Unity® Exome Plus Analysis | Genomic Unity® Exome Analysis | Genomic Unity® Constitutional Genome-Wide Copy Number Variant Analysis | Genomic Unity® Genome-Wide CNV and FMR1 Analysis | Genomic Unity® Neuromuscular Disorders Analysis | Genomic Unity® Muscular Dystrophy Analysis | Genomic Unity® DMD Analysis
Want similar results for your patients?
Connect with a Clinical Specialist to find out how easy it is to bring the power of whole genome sequencing into your practice.
