MISSED BY OTHERS, DETECTED BY US
Genomic Unity® Case Study
Clinical presentation
A 53-year-old male with a clinical diagnosis of Diamond-Blackfan
anemia (in remission for 26 years) presented with the following symptoms:
- Short stature
- Dyspnea, history of myocardial infarction
- History of kidney stones
- Blindness
- Hearing impairment
Previous genetic testing
Multiple tests were performed with negative results including:
- Diamond-Blackfan anemia panel
- Bone marrow failure disorders panel
- Whole exome sequencing (WES)
Genomic Unity® Testing
was ordered because of its ability to identify all major variant types in a single test.
Results and interpretation
Variantyx Genomic Unity® testing identified a heterozygous, pathogenic deletion of 811-818 kb in the chr3q29 region.
The deletion spans the entire RPL35A gene.
Diagnosis: Diamond-Blackfan anemia
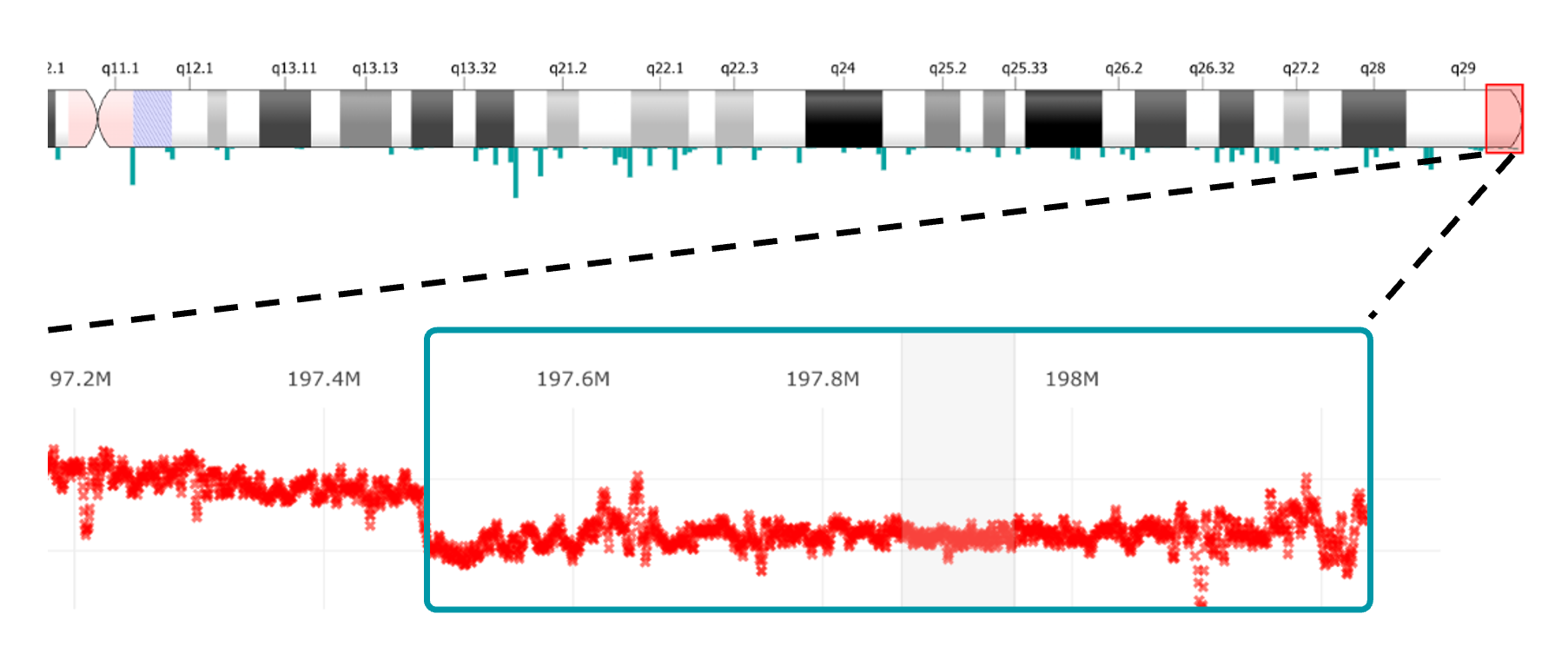
Uniform data from WGS clearly shows that the chr3q29 region has half the expected variant allele frequency compared to surrounding regions, indicative of a heterozygous deletion.
The Variantyx Difference
Why was this heterozygous, full gene deletion detected by Genomic Unity® testing, and not detected by prior testing – including panel and exome testing?
-
This large deletion could have been detected by both panel and exome testing. However, the provider’s analysis tools may not have been sensitive enough to detect changes in read coverage, missing the deletion.
Variantyx genome sequencing’s uniform coverage depth across the entire genome makes it easy to identify this heterozygous, full gene deletion. -
The breakpoint of this terminal deletion falls in an intergenic region, adding to the complexity of detection.
Variantyx genome sequencing includes intronic and intergenic regions, enabling breakpoint detection regardless of location.
Variantyx tests that would have identified this variant
Want similar results for your patients?
Connect with a Clinical Specialist to find out how easy it is to bring the power of whole genome sequencing into your practice.
