MISSED BY OTHERS, DETECTED BY US
IriSight® Case Study
Clinical presentation
A multigravida underwent an anatomy scan at 22 weeks gestation. In addition to IUGR and minimal fetal movement, the following fetal findings were noted:
- Non-immune hydrops, fetal ascites, pleural effusion
- Hand clenching, club foot, rocker bottom feet
- Short long bone
- Arthrogryposis multiplex congenita
Previous genetic testing
Testing was performed with negative results including:
- Chromosomal microarray
- Whole exome sequencing
IriSight® Testing
was ordered because of its ability to identify all major variant types in a single test.
Results and interpretation
Variantyx IriSight® testing identified a homozygous, pathogenic, maternally and paternally inherited 13.13 kb deletion encompassing DOK7 exon 7.
Diagnosis: DOK7-related disorders
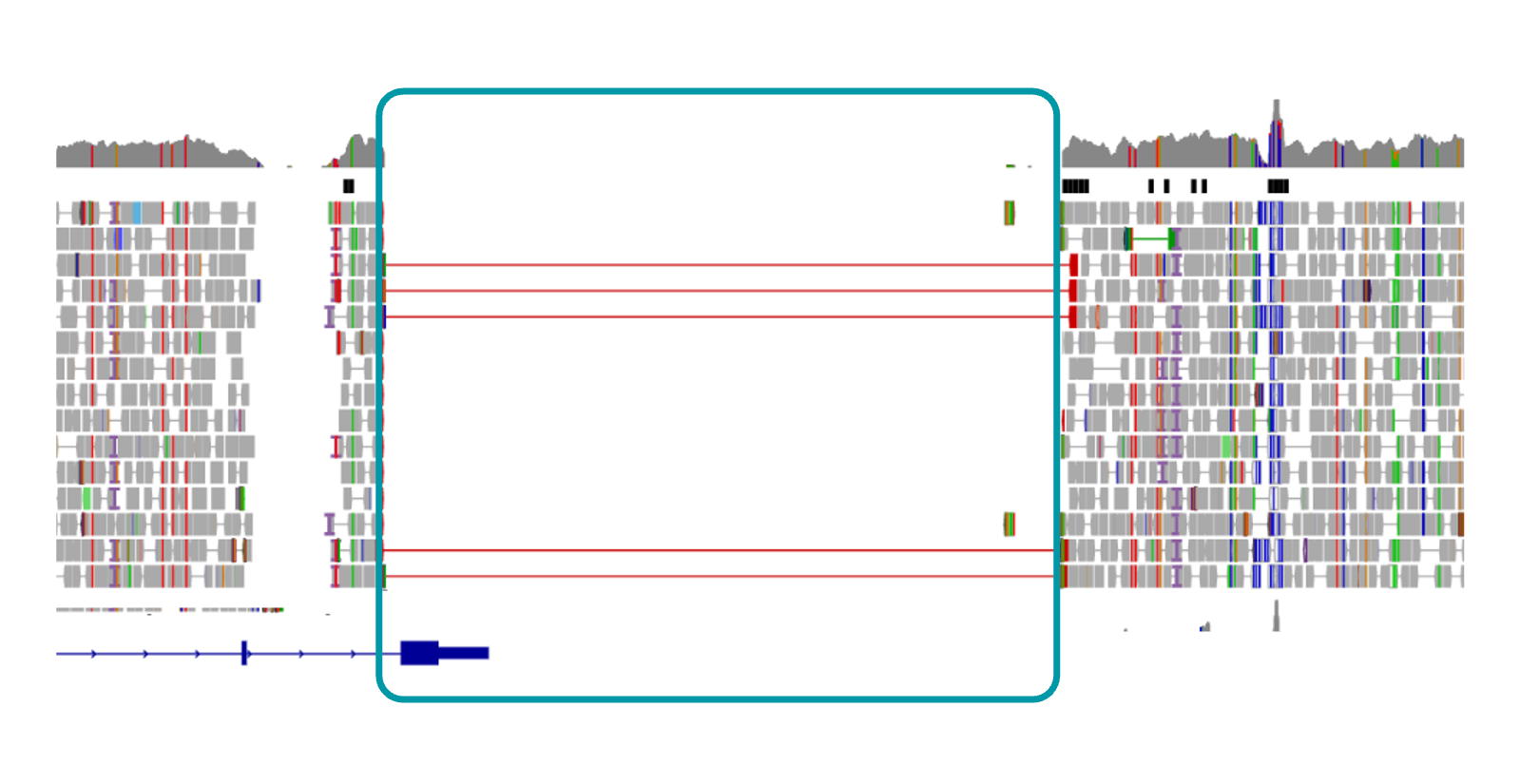
Uniform data from WGS clearly shows the single exon DOK7 deletion.
The Variantyx Difference
Why were these variants detected by IriSight® testing, and not detected by chromosomal microarray or exome testing?
-
The size of the 13.13 kb deletion falls below the cut-off value for CMA (~25 kb).
-
Exomes are typically unable to detect deletions smaller than 3 exons in size. The exome report specifically noted that CNV analysis was attempted using the sequencing data but could not be completed.
Variantyx genome analysis has a detection range from 1bp to whole chromosomal events, easily detecting the single exon deletion.
Want similar results for your patients?
Connect with a Clinical Specialist to find out how easy it is to bring the power of whole genome sequencing into your practice.
