
MISSED BY OTHERS, DETECTED BY US
Genomic Unity® Case Study
Clinical presentation
A 15-year-old male of consanguineous parents presented with clinical suspicion of a progressive neuromuscular disorder, plus the following symptoms:
- Chronic respiratory disease
- Chronic nocturnal hypoventilation
- Hypoxemia
- Ophthalmoplegia
Previous genetic testing
Testing was performed with non-diagnostic results. Including exome testing that identified a likely pathogenic SNV in the OPA1 gene, explaining only the ophthalmoplegia symptom.
- Whole exome sequencing

Genomic Unity® Testing
was ordered because of its ability to identify all major variant types in a single test.
Results and interpretation
Variantyx Genomic Unity® testing identified a homozygous, pathogenic, 2.77 kb single exon deletion in the COLQ gene together with a homozygous, likely pathogenic SNV in the CFTR gene – providing two independent molecular diagnoses.
The likely pathogenic SNV in the OPA1 gene was also identified.
Diagnosis: Congenital myasthenic syndrome 5, Cystic fibrosis, Optic atrophy
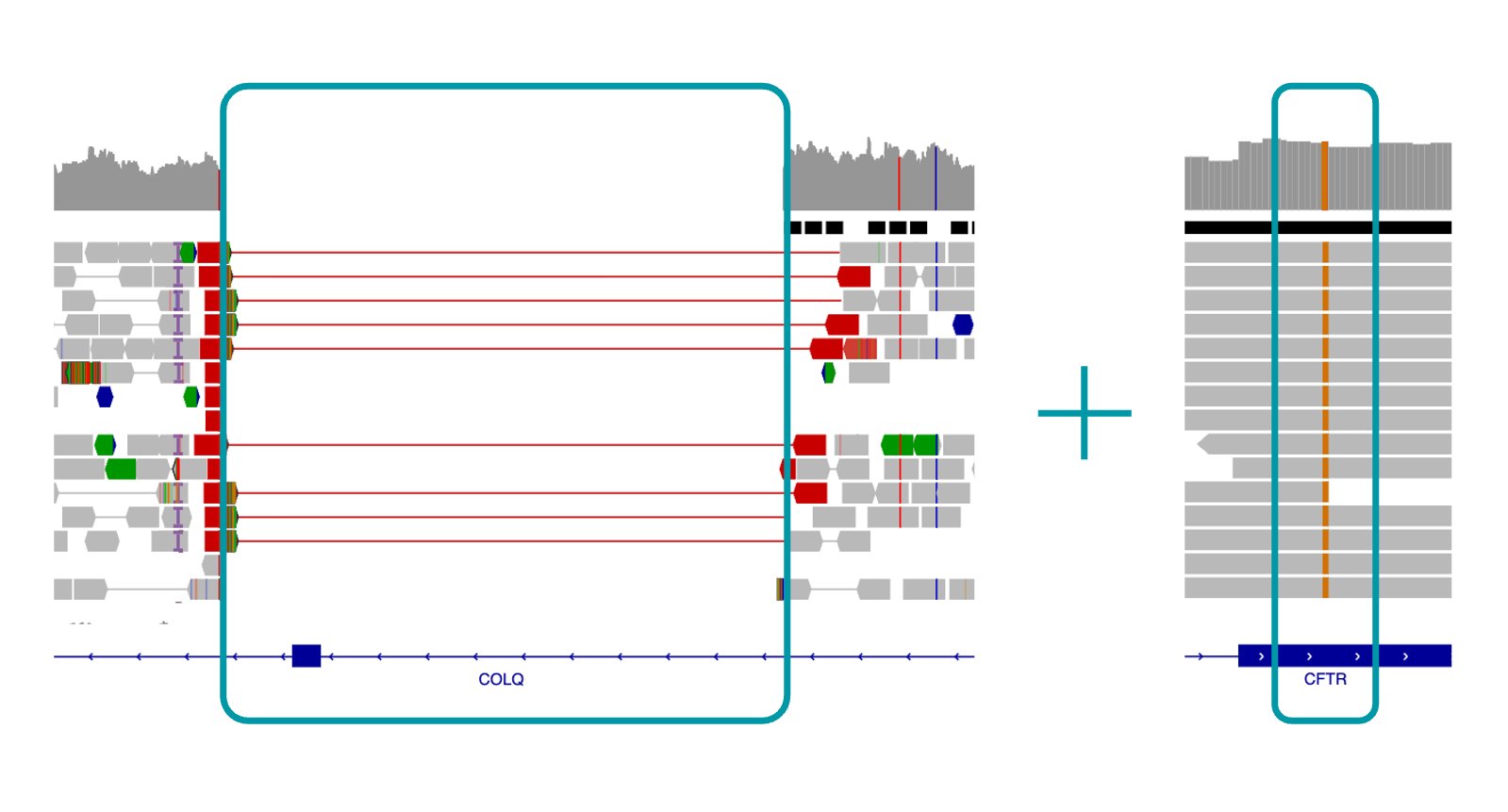
Uniform data from WGS clearly shows both the 2.77 kb COLQ deletion (left) and CFTR SNV variant (right) inherited from both parents. The maternally inherited OPA1 SNV is not shown.
The Variantyx Difference
Why were these variants detected by Genomic Unity® testing, and not detected by exome testing?
-
Exomes are typically unable to detect deletions smaller than 3 exons in size.
Variantyx genome analysis has a detection range from 1 bp to whole chromosomal events, easily detecting this single-exon COLQ deletion. -
Both COLQ deletion breakpoints are intronic, adding to the complexity of detection.
Variantyx genome analysis includes intronic regions, enabling breakpoint detection regardless of location. -
While small sequence changes are usually detectable by exome testing, this CFTR variant is located near an exon/intron boundary that may have impacted probe coverage and ultimately read coverage.
Variantyx genome sequencing does not use PCR amplification. Its uniform coverage across the entire gene, including at exon/intron boundaries, makes it easy to identify this exonic single nucleotide change.
Variantyx tests that would have identified this variant
Want similar results for your patients?
Connect with a Clinical Specialist to find out how easy it is to bring the power of whole genome sequencing into your practice.
